Gene Knockout Validation
Introduction:
CRISPR/Cas9 is the third generation of gene editing technology following Zinc Finger Nuclease (ZFN) and Transcription-Activator Like Effector Nucleases (TALEN). This technology has been developing rapidly since 2013. In the CRISPR Cas9 system, a sgRNA directs the Cas9 endonuclease to the target loci and then cleave the target gene. Since the gene editating efficiency of the gRNA is unpredictable, therefore, the choice of gRNA with high efficiency and specificity is critical to successful gene editating. For the efficient gRNA screening, researchers typically test multiple gRNAs(3 to 5) per target gene.
Service Details
WZ Biosciences can provide personalized Gene Kockout Validation Services for you according to your research needs, including Custom gRNA Validation and efficient gRNA Validation.
|
Cat No.
|
Type
|
Validation Method
|
Price
|
Turnaround Time
|
|
WZ100001-1
|
Custom gRNA Validation
|
Fluorescence Method
Cloning and Sequencing
|
$1500/test
|
1-2weeks
|
|
WZ100001-2
|
Custom gRNA Validation
|
PCR Products Sequencing
|
$1000/test
|
1-2weeks
|
|
WZ100002-1
|
Efficient gRNA Validation
|
Fluorescence Method
Cloning and Sequencing
|
$1500/test
|
1-2weeks
|
|
WZ100002-2
|
Efficient gRNA Validation
|
PCR Products Sequencing
|
$1000/test
|
1-2weeks
|
*Interested in above Kockout Validation Services? Please feel free to contact us to“Request a quote”.
Related services: CRISPR/Cas9 Cloning, Adenovirus Packaging, Lentivirus Packaging, AAV Packaging, Stable Cell Line Generation
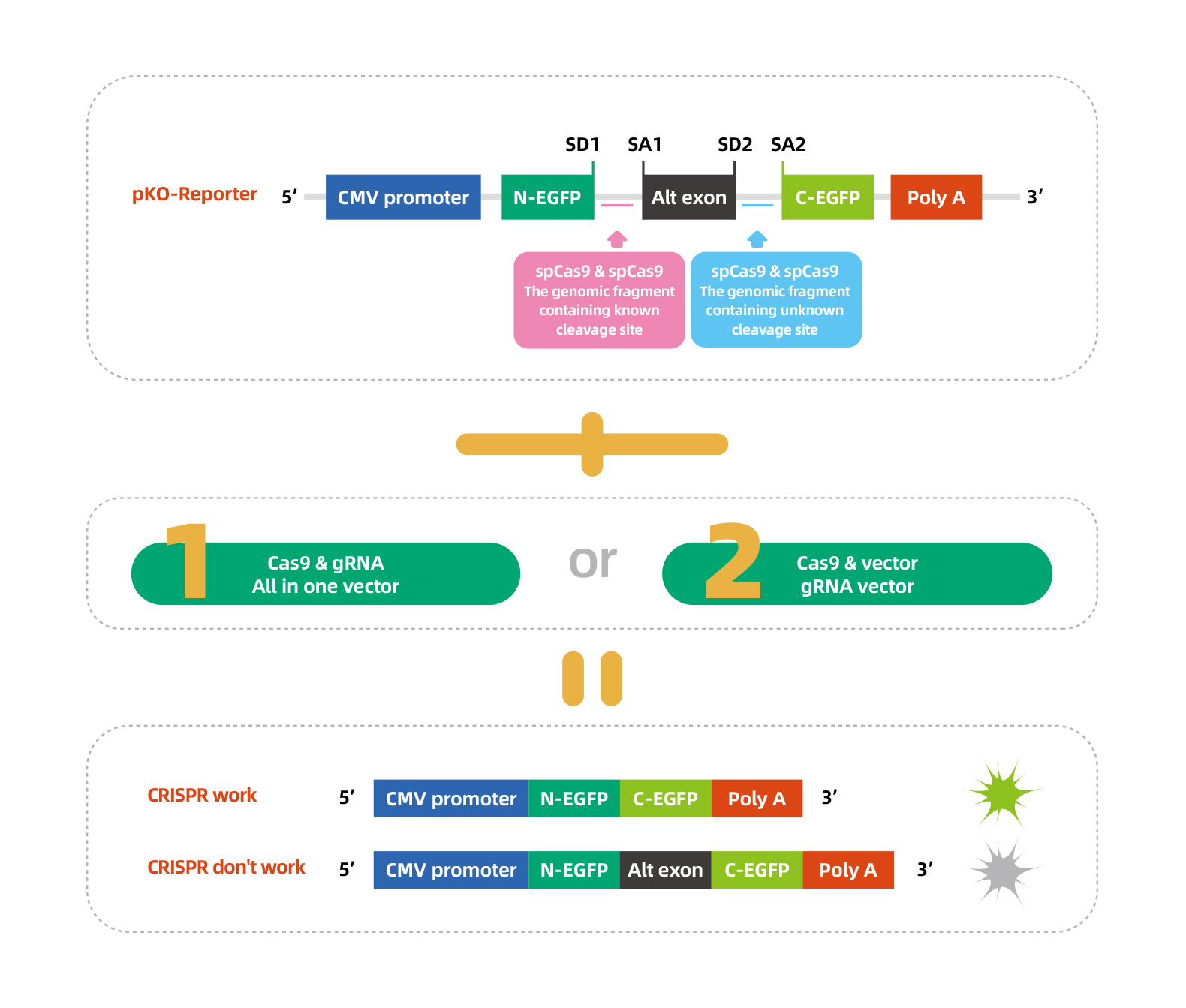
Fluorescence Method
In CRISPR/Cas9 experiments, researchers often use sequencing methods to confirm gene edits, including sanger sequencing of PCR products(qualitative) and sanger sequencing of cloned inserts(quantitative). Even though the results of these two methods are reliable, they have several limits, such as complex operation, long-term experiment, cell types limitation, and so on. To address these problems, WZ Biosciences have developed a simple validation system-Fluorescence Method.
Design Strategy
|
|
Advantages:
|
|
|
|
1、Easy-to-use;
2、Short cycle, usually completed within 3 working days;
3、Confirmation of genome editing is not limited by cell types;
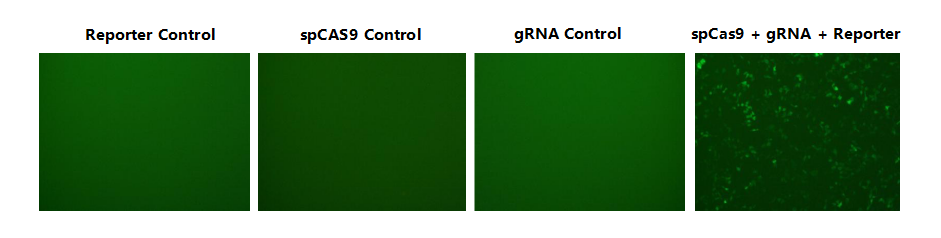
4、The knockout effect is visible. The knockout effect is detected by whether EGFP emits light or not; the knockout efficiency is qualitatively determined by the luminous intensity of EGFP.
5、The experimental results is reliable, the experimental data are consistent in batches.
|
|
Advantages:
Multiple gRNAs targeting some gene are designed and construted. Then we co-transfect the pKO-Reporter plasmids and gRNA & Cas9 expression plasmids into HEK293 cells. Validation of shRNA knockdown efficiency was assessed by the GFP reporter gene. Images below were taken 48h post-transfection.
The chart below shows the comparsion between sanger sequencing of PCR products, sanger sequencing of cloned inserts and Fluorescence Method:
|
Validation Method
|
Validation Method
|
qualitative/quantitative
|
Accuracy
|
cell types limitation
|
|
sanger sequencing of PCR products
|
Short,4-5 working days
|
qualitative,Gene editating efficiency is validated by observing sequencing results
|
High,Site-specific cleavage of genomic DNA
|
Yes
|
|
sanger sequencing of cloned inserts
|
Long,10-15working days
|
quantitative,Gene editating efficiency is validated by observing sequencing results
|
High,Site-specific cleavage of genomic DNA
|
Yes
|
|
Fluorescence Method
|
Short,3 working days
|
qualitative,Gene editating efficiency is validated by observing EGFP fluorescence
|
High,The genomic fragment containing the sgRNA target sequence is cloned into a reporter vector
|
Yes
|
Service Process
Related Resorces

|
《Transfection Reagent Instructions》
|
|
|
|
|
|
|
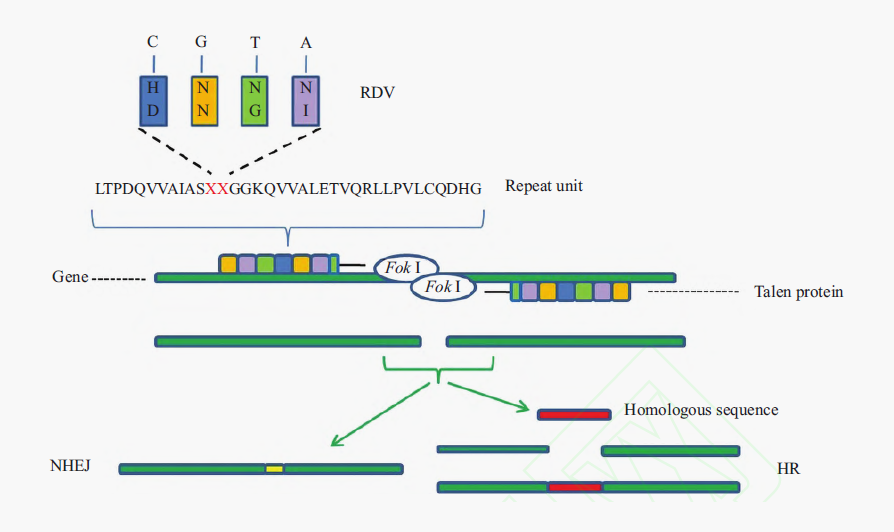
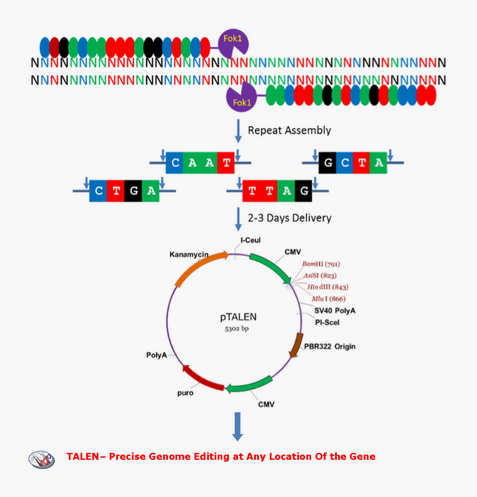
Transcription Activator-like Effector Nucleases (TALENs) are artificial restriction enzyme that can cut specific sequences of DNA. TALENs are designed by fusing a TAL effector DNA-binding domain to a DNA cleavage domain (a nuclease), which cuts DNA strands. TALENs introduce a site-specific double-strand break (DSB), which is subsequently repaired by the endogenous DNA repair machinery, such as non-homologous end-joining (NHEJ) and homology-directed repair (HDR), which often creates short DNA insertions or deletions (indels) . The principle is shown in the figure below:
TALEs (TAL effectors,Transcription activator-like effectors)
The TAL effector (TALE) is natural protein secreted by plant pathogenic bacteria Xanthomonas sp., which can recognize specific DNA base pairs.
Design Strategy
|
|
Advantages:
|
|
|
|
1.A complete collection of repeat tetramers for assembling 16-20 repeat units directly;
2.Various virus vectors available;
|
3. Both clones are Fully sequenced;
4. Functional validation available。
|
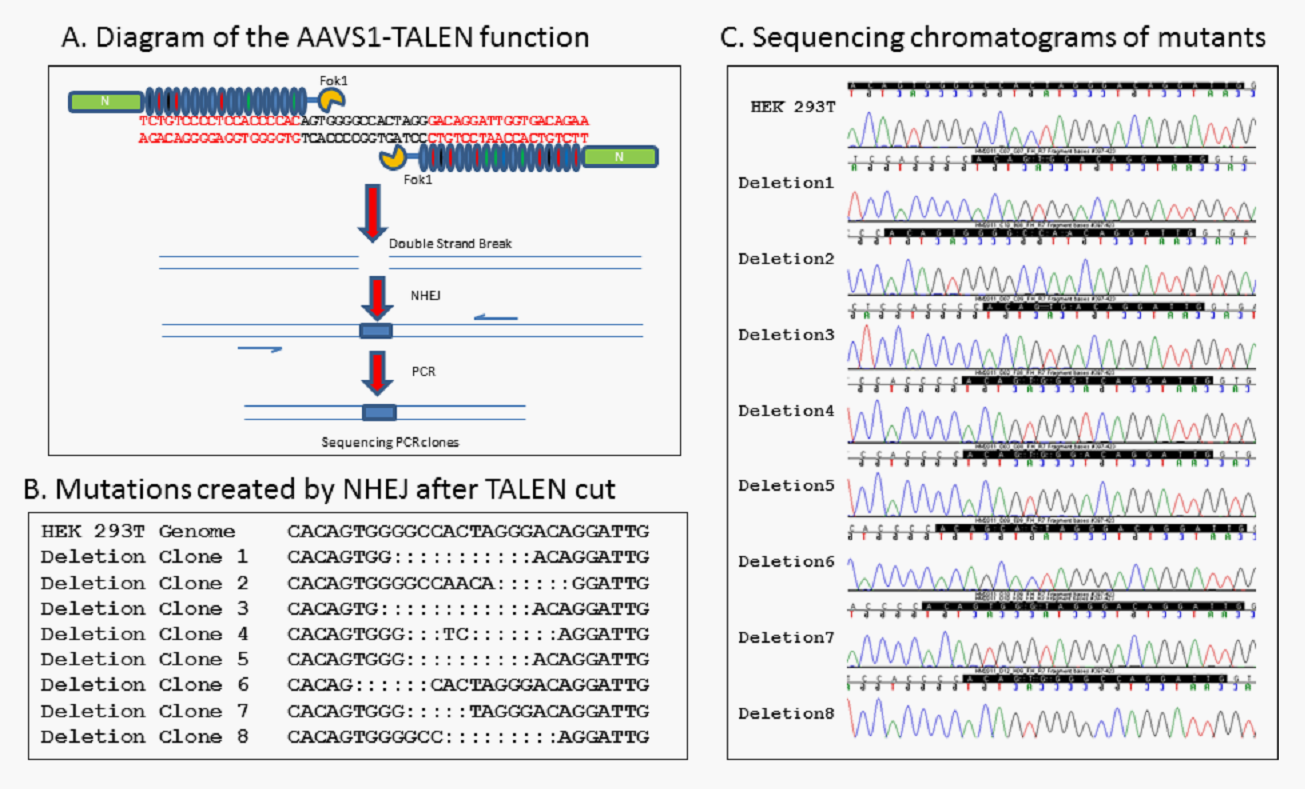
Application example---- Integration site AAVS1 gene editing in human genome by AAV transfection
Two(Left & Right) AAVS1-TALEN were created using WZ Biosciences’ Custom TALEN Design(Fig.A). AAVS1-TALEN was then sequenced to confirm for TALEN cut outcom(Fig.B & C).
Results: 15 clones were sequenced, 9 contains deletions with 8 being different deletions, results in 8 mutations created by the TALEN design.
|
|
Comparison of TANLEN & CRISPR/Cas9
|
Property
|
TALEN
|
CRISPR-CAS9
|
|
Type of recognition
|
Protein-DNA
|
RNA-DNA
|
|
Modularity
|
One RVD recognizes one nucletide
|
one nucletide to one nucletide
|
|
Target sequences
|
2×16 nucletide and up
|
~20 nucletide
|
|
Off-target effect
|
Less observed off-target effects
|
More potential off-target effects
|
|
Multiplexing
|
Rarely used
|
Capable
|
|
Methylation sensitive
|
Sensitive
|
Not sensitive
|