CRISPR/CasRx cloning
The CRISPR/Cas13 system is currently the only protein discovered that can degrade RNA, and its discovery extends the application of the CRISPR system in virus interference. The CRISPR/CasRx system is a family member of the newly discovered fourth subtype of Cas13 protein (Cas13d), which exhibits stronger specificity and higher knockout efficiency in RNA cleavage processes in mammalian cells both in vivo and in vitro. Moreover, CasRx is much smaller than all previously discovered subtypes, making it easier to transport in vivo using AAV vectors. Therefore, it has great potential for application in RNA editing.
Weizhen Biotechnology provides CasRx AAV vector (see service content below), which has higher gene editing efficiency and better results. Welcome to exchange and order with teachers!
Service Number
|
Service Type
|
Specifications
|
Catalog price
|
Delivery time
|
WZ140001
|
a single gRNA plasmid construct
|
5ug DNA;500ul Glycerol Stock
|
inquiry
|
inquiry
|
WZ140002
|
2-in-1gRNA plasmid construct
|
5ug DNA;500ul Glycerol Stock
|
inquiry
|
inquiry
|
WZ140003
|
CasRx plasmid construct
|
5ug DNA;500ul Glycerol Stock
|
inquiry
|
inquiry
|
Service Features
1、The gene knockout effect is good.
Partial CasRx Adeno-Associated Virus (AAV) Vector
|
|
pAV-CMV-nls-CasRx pAV-CMV-nls-CasRx-P2A-GFP
pAV-U6-gRNA-CMV-nls-CasRx pAV-U6-gRNA-CMV-nls-CasRx-P2A-GFP
|
Experimental Test Results
The Weizhen Research Team designed multiple gRNAs targeting the mcherry gene and selected two gRNAs (gRNA1 and gRNA2) with good knockout effects for validation experiments of single gRNA and 2in1 gRNAs knockout effects, as follows:

From the above fluorescence images, it can be seen that in the 2in1 gRNAs knockout group, the expression level of mCherry is significantly reduced, and the knockout effect is much better than that of single gRNA knockout.
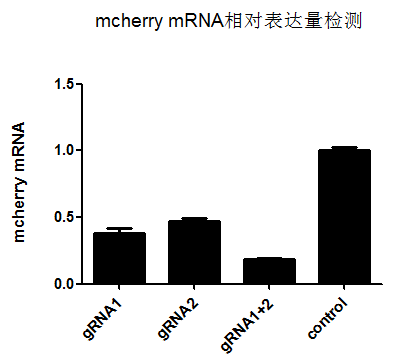
(mCherry expression level: gRNA1 group 0.38; gRNA2 group 0.47; gRNA1+2 group 0.18)
In addition, from the experimental results of mCherry mRNA expression detection in the above figure, it can be seen that the Cas13d (CasRx) - gRNA system can specifically and efficiently reduce the expression of target genes (mCherry), and the 2-in-1 gRNAs can achieve a 82% knockdown at the mRNA level!
2. Complete carrier
Using AAV vectors to construct clones, over 40 tissue-specific promoters and multiple reporter genes are available for customers to choose from.
3. Excellent service
Based on specific customer experiments, the company's professional technical personnel can provide carrier construction solutions and offer special customized services.
4. Fair and reasonable prices
Common carriers
1、Introduction to CRISPR/Cas13 System
In 2016, scientists discovered the CRISPR/Cas13 system that can target RNA for cleavage. This system consists of a single effector protein Cas13 and CRISPR RNA (crRNA) assembled to form an RNA targeted effector complex guided by crRNA. At present, four subtypes of the Cas13 family have been identified, including Cas13a (also known as C2c2), Cas13b, Cas13c, and Cas13d, all of which have protein molecular weights smaller than Cas9 protein.
All Cas13 proteins have two different catalytic activities:
① The RNase activity provided by two higher eukaryotic and prokaryotic nucleotide binding (HEPN) domains is essential for the degradation of target RNA. The homology of the four subtypes of Cas13 is low, and the homologous sequences are limited to the HEPN domain sites. In addition, single point mutations in two HEPN domains can completely impair the CRISPR/Cas13 system's ability to cleave RNA;
② The RNase activity that catalyzes pre crRNA processing and the formation of mature crRNA.
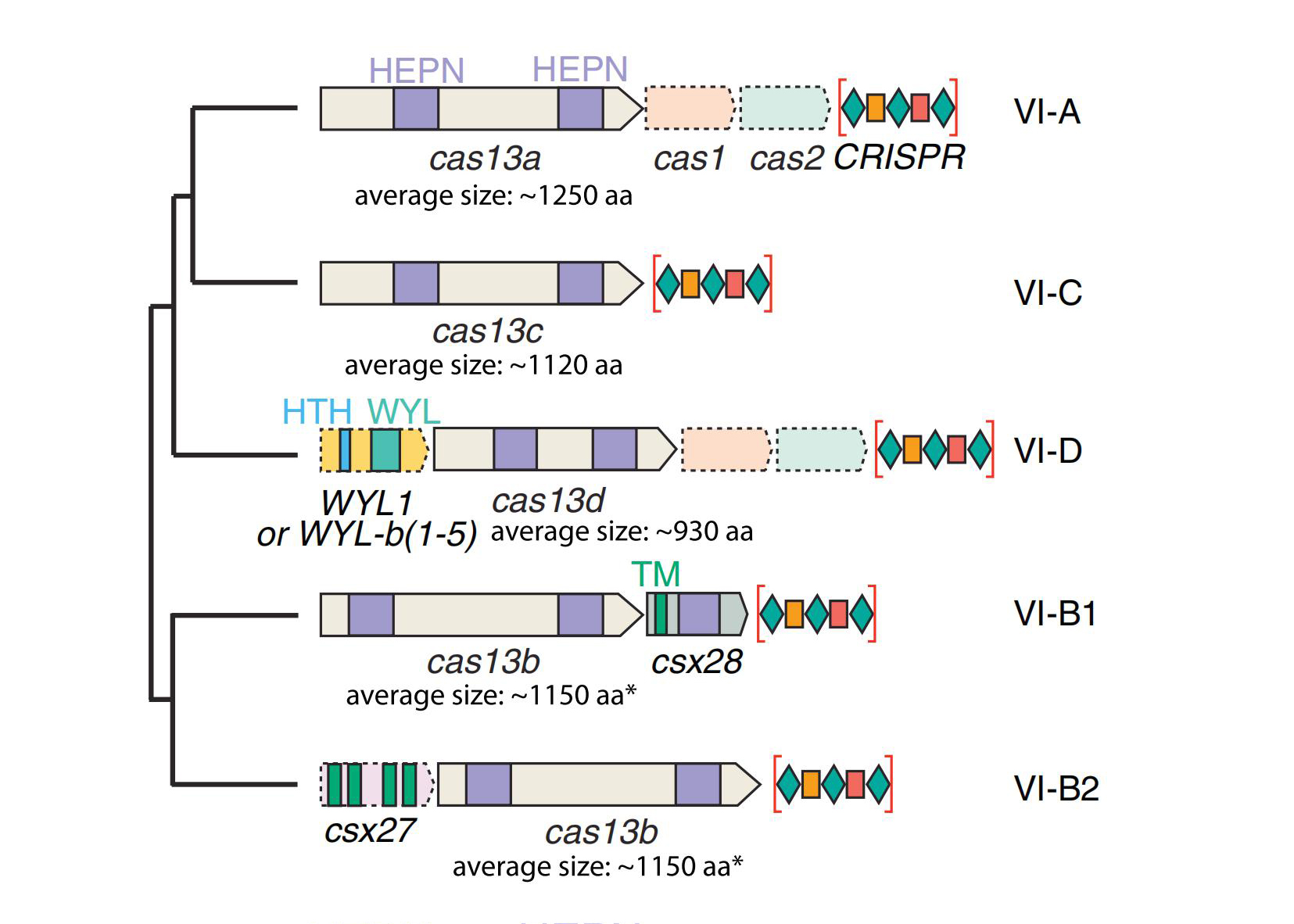
Figure 1. Introduction to CRISPR/Cas13 system
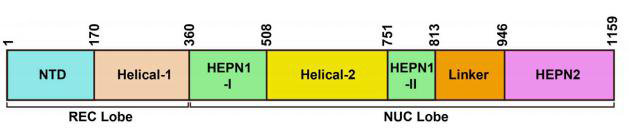
Similar to other nucleases in the CRISPR class II system, the overall structure of Cas13a is biplate, with the N-terminal domain (NTD) and Helical-1 domain forming the crRNA recognition lobe (REC lobe), and the HEPN1, Helical-2, and HEPN2 domains forming the nuclease lobe (NUC lobe) (Figure 2). The CRISPR/Cas13b system has two enzymes, VI-B1 and VI-B2. The difference between them is the genotype of the accessory protein carried on the Cas13b transposon. The accessory protein of VI-B1 is Csx28, while the accessory protein of VI-B2 is Csx27. The vast majority of Cas13d type systems also contain auxiliary proteins containing WYL domains, which are typically associated with prokaryotic defense systems (Figure 1).
Figure 2. Schematic diagram of Cas13a protein domain
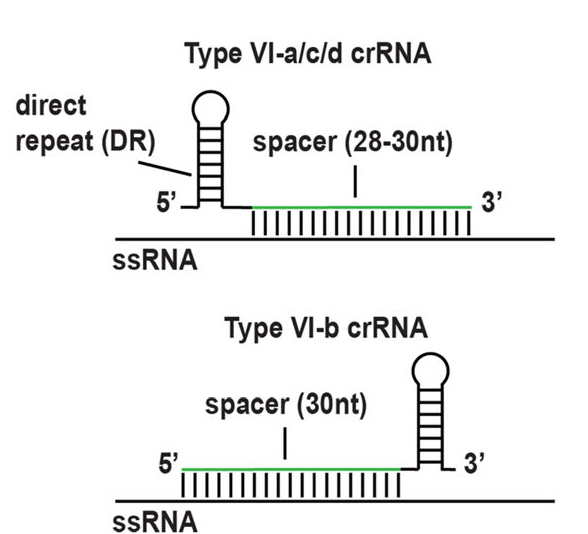
All Cas13 proteases require 60-66 nt crRNA to ensure targeting specificity. All four subtypes of crRNA have a direct repeat (DR) that promotes binding to the corresponding Cas13 protease and a spacer sequence that specifically targets the transcript. The crRNA of Cas13b carries a direct repeat at the 3 'end, while the crRNAs of Cas13a, c, and d carry a direct repeat at the 5' end (Figure 3).
Figure 3. Schematic diagram of Cas13a protein domain
2、The defense mechanism of CRISPR/Cas13 system
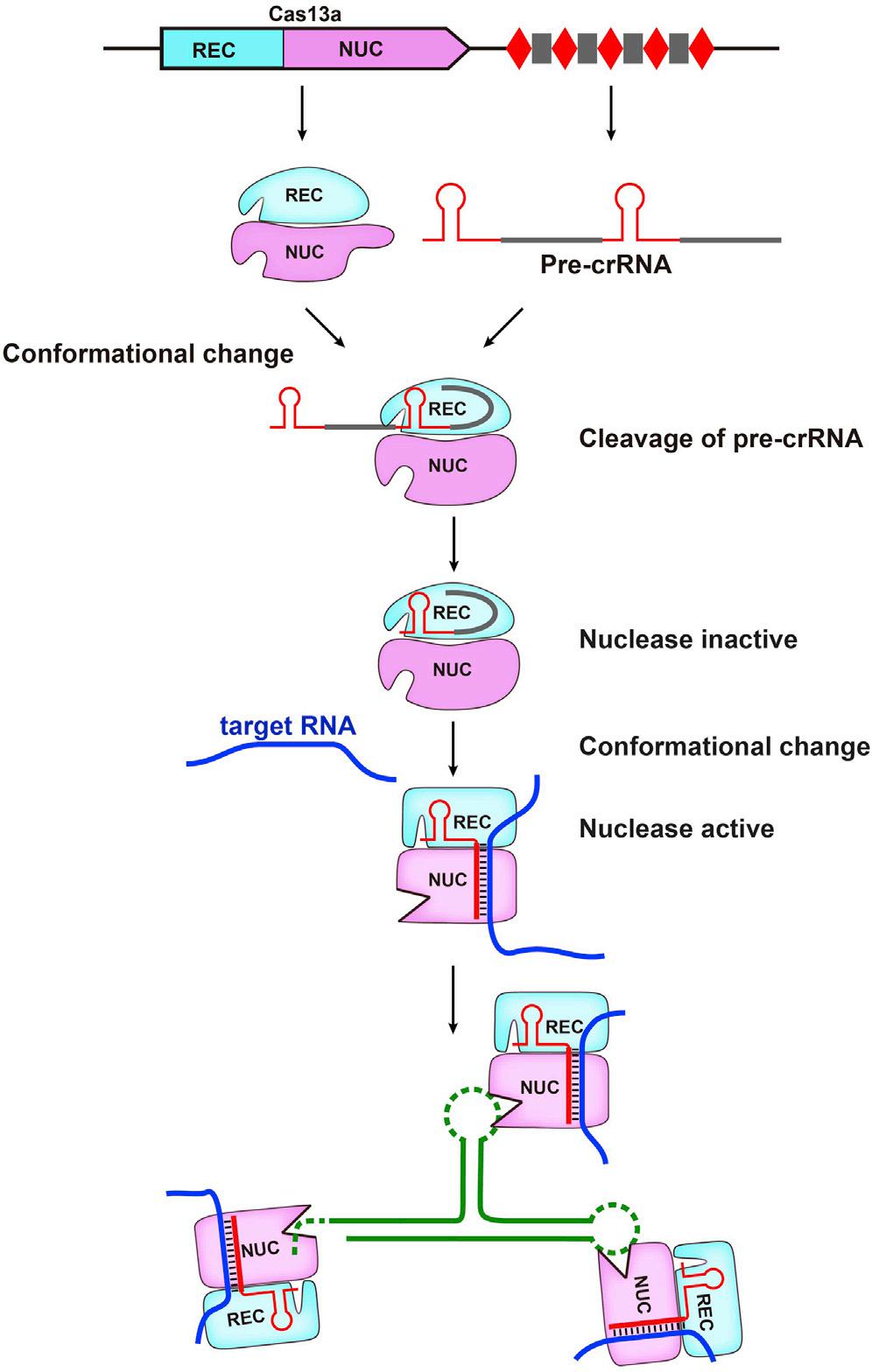
It mainly includes the following stages (taking Cas13a as an example):
① Recognition and binding of pre crRNA. The newly transcribed pre crRNA recognizes and binds to the REC leaf of Cas13a through the 5 'end stem loop structure of crRNA, forming an intermediate transition state of the pre crRNA Cas13a complex;
② The formation of mature crRNA. The conformation of conserved residues between the Helicol-1 and HEPN2 domains in the NUC region undergoes a change, forming an acid-base catalytic center that catalyzes enzyme cleavage of pre crRNA to form mature crRNA. At this point, the crRNA Cas13a complex is in a state of no enzymatic activity;
③ Activation of crRNA-Cas13 complex enzyme cleavage activity. The target ssRNA enters the crRNA Cas13a complex and undergoes base pairing with crRNA, inducing conformational changes in Cas13a and activating the enzymatic activity of crRNA Cas13a complex;
④ Degradation of target RNA. Under the guidance of crRNA, the HEPN domain of Cas13a catalyzes the enzymatic cleavage of target ssRNA. Sometimes non-specific enzyme cleavage occurs in bacterial cells, leading to the degradation of other accessory ssRNAs in the cell and causing certain cytotoxicity. However, this phenomenon has not occurred in mammalian cells, and the reason for it is currently unknown. (Figure 4).
Figure 4. Schematic diagram of the mechanism of action of CRISPR/Cas13 system
3、Discovery and advantages of Cas13d
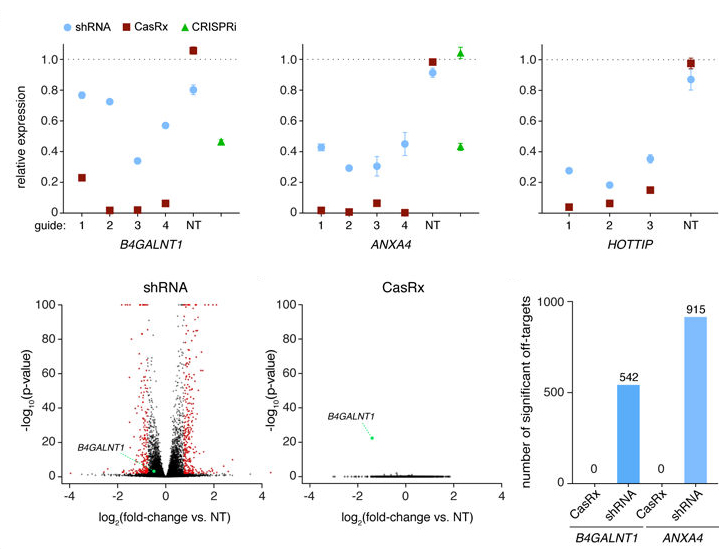
In 2018, Patrick D. Hsu's laboratory at the University of California, Berkeley discovered the Cas13d family, which targets RNA. Cas13d has an average size of 930 aa and is currently one of the relatively small class II CRISPR effectors found in mammalian cells (20% smaller than its family members Cas3a-c and 33% smaller than Cas9), making it easier to package into limited capacity application vectors such as AAV vectors. In addition, the cleavage of RNA targets by Cas13d does not rely on the PFS sequence (protospacer flanging sequence, equivalent to Cas9's PAM sequence for DNA), which greatly increases the application scope of Cas13d and makes it a potential platform for further development of targeted RNA tools. Compared with RNA interference technology, Cas13d mediated gene silencing has higher specificity (compared to hundreds of shRNA off targets, Cas13d did not off target) and knockout efficiency (Cas13d reached 96%, shRNA reached 65%, CRISPRi reaches 53%. Compared with Cas9 mediated gene knockout technology, Cas13d mediated gene silencing does not alter genomic DNA, making this gene silencing reversible and more advantageous for the treatment of some acquired diseases (Figure 5, 6).
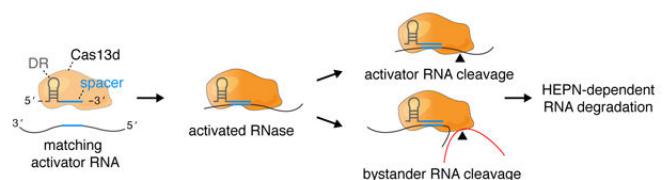
Figure 5. A model for guiding and targeting the activation of Cas13d ribonuclease activity
Figure 6. Cas13d system has higher specificity and knockout efficiency
4、Verification of the Knockout Effect of CasRx
▽ In vitro experiments:
A 2020 study explored the feasibility of using the CasRx system to selectively silence genes. Firstly, the researchers chose Pten as a metabolic regulatory gene to investigate whether CasRx can effectively knock out genes targeting metabolic genes. Multiple sgRNAs targeting Pten mRNA coding sequences were prepared, and CasRx and each Pten sgRNA were transduced into mouse neuroblastoma N2a cells through plasmids. In vitro screening of sgRNAs was conducted, and the optimal sgRNA combination for CasRx knockdown of the target gene was found: sgPten-5 and sgPten-6. The results showed that in N2a cells transfected with CasRx, sgPten-5, and sgPten-6, the expression level of Pten was significantly reduced, and the phosphorylation level of AKT was significantly increased. In addition, compared with the shRNA system, the CasRx editing system exhibited stronger specificity and lower off target rate (Figure 7). (Extended understanding: Pten is considered a metabolic regulator that inhibits the insulin signaling pathway by suppressing the PI3K/AKT pathway. The absence of Pten can promote AKT phosphorylation.)
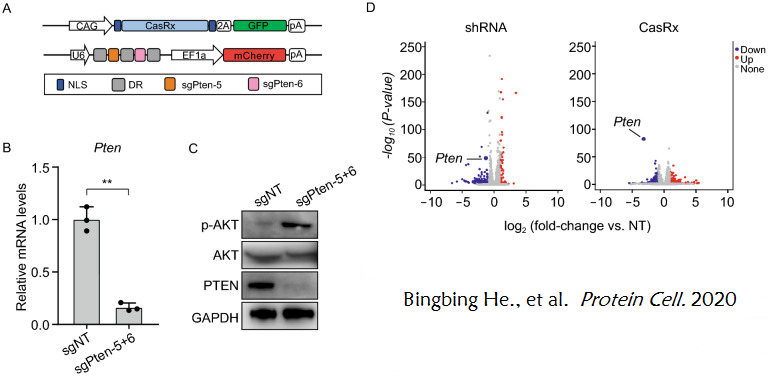
Figure 7. CasRx mediated Pten knockout in vitro
▽ In vivo experiment:
Researchers injected AAV8 virus knocking down Pcsk9 through the tail vein, significantly reducing the expression levels of Pcsk9 in the liver and serum, as well as cholesterol levels in the serum. Importantly, CasRx mediated knockdown of Pcsk9 is reversible, and Pcsk9 expression levels can be repeatedly downregulated. The CRISPR/CasRx system provides an effective strategy for reversible regulation of metabolic genes, especially in some acquired diseases such as hyperlipidemia caused by unhealthy lifestyle habits (Figure 8).
Figure 8. CasRx mediated serum cholesterol reduction and PCSK9 reversible regulation